Surveillance of Influenza A virus in Danish pigs
On this webpage the results of the national swine influenza A virus surveillance will continuously be updated.
The results of the swine influenza A virus surveillance will continuously be updated on this webpage with a monthly frequency. The results include a brief overview and relevant figures.
As all data is not always available by the end of a given month, there can be slight delays in the update of the results.
For previous monthly results and annual reports please navigate to ”Archive”.
Latest results
Samples and results 2025
The table illustrate the number of samples, submissions and herds that contributed to the surveillance program each month. In addition, the results of the influenza A virus and H1pdm09 screening are shown.
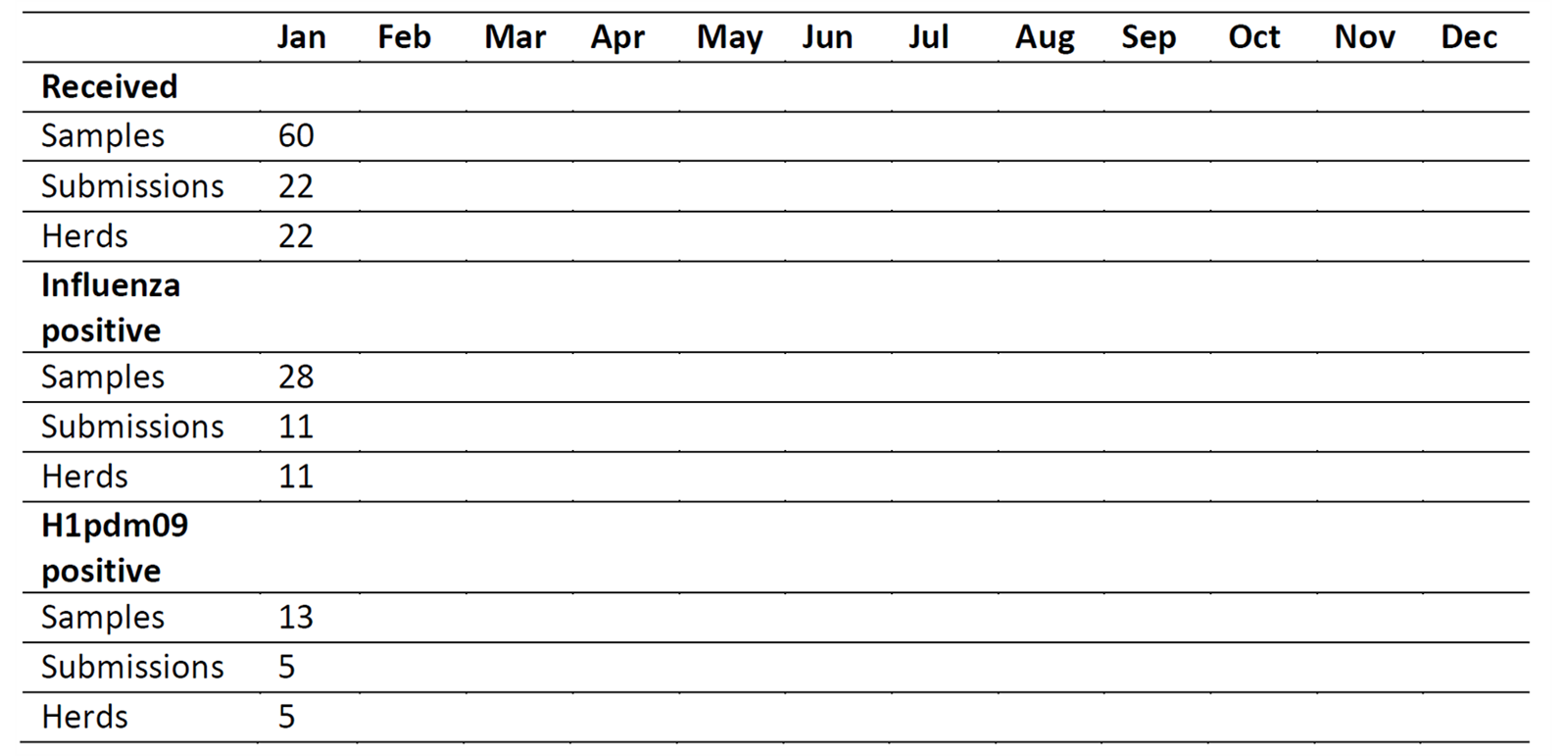
In December, 58 submissions from 58 herds registered with different CHR numbers were received. On average, each submission consisted of 3.4 samples. The percentage of influenza positive submissions was 60% resembling the level observed in August and September. All influenza A virus-positive samples were tested for the presence of H1pdm09. Overall, 40% of the submissions testing positive for the influenza A virus were positive for H1pdm09. 
The figure illustrates the percentage of influenza A virus negative and positive submissions including the proportion of H1pdm09 positive submissions. The share of submissions with H1pdm was 35%, which is comparable to previous months earlier in the year.
Distribution of swine influenza A virus subtypes
Swine influenza A virus can be classified into subtypes and genotypes. The subtype describes the combination of HA and NA surface gene segments, and the genotype describes the combination of all eight genome segments based on their genetic origin. Information on the contemporary circulating swine influenza A virus subtypes is essential for the update of vaccination protocols, optimization of the diagnostic assays and for evaluation of the zoonotic risk.
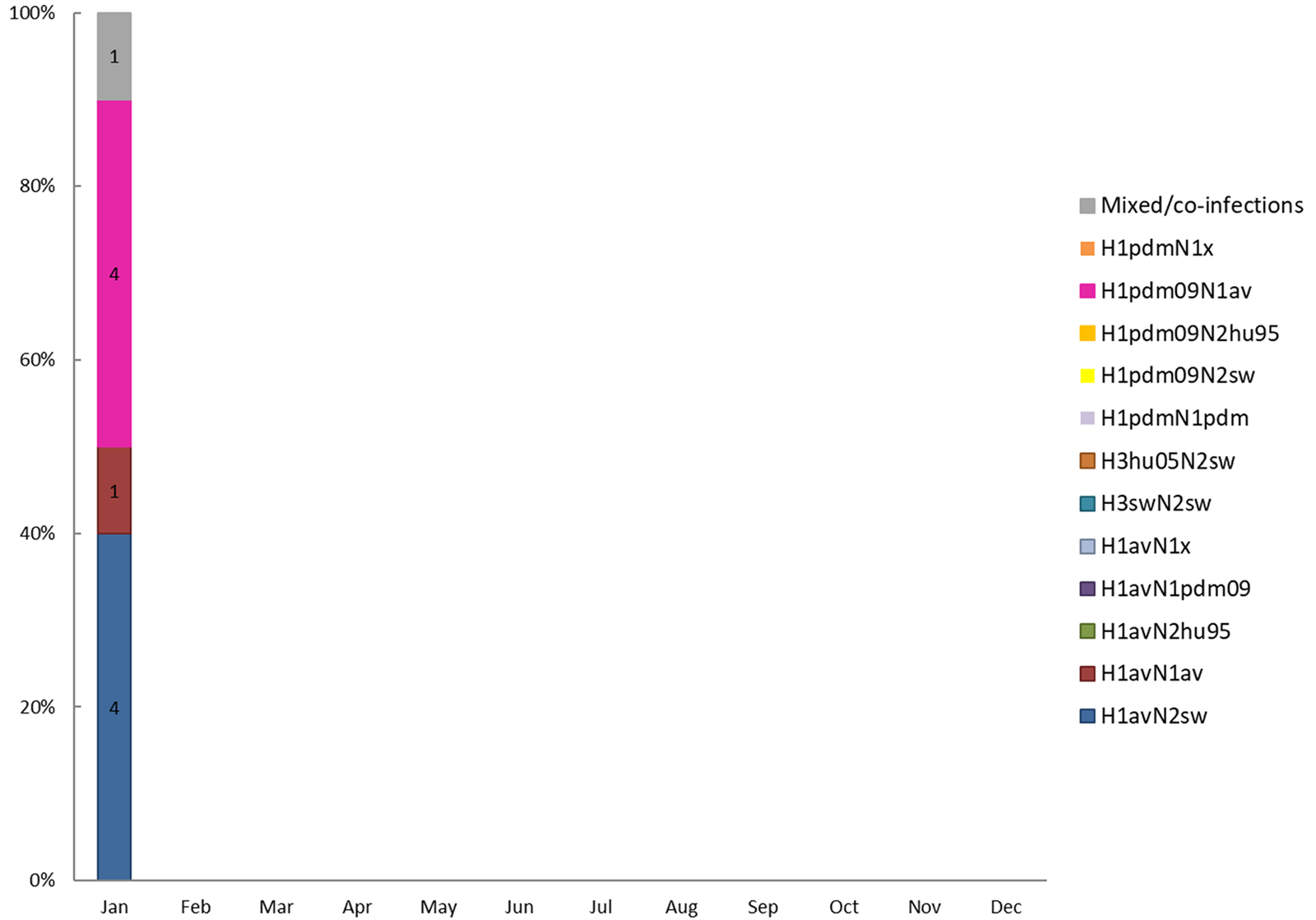
In December, the full subtype (both HA and NA gene segments) was identified for 25 submissions. The majority were of the H1avN2sw subtype (n=12) while the H1pdm09N1av was observed in five submissions. In addition, H1pdmN1pdm and H1pdmN2sw were observed in three and two submissions, respectively. H1avN2hu95 was observed in a single submission for the first time since 2022. In addition, a submission was positive for H1avN2x carrying a novel N2 gene, that remains to be fully classified. This month one submission showed signs of co-infections with several swIAV strains.
Distribution of swine influenza A virus genotypes
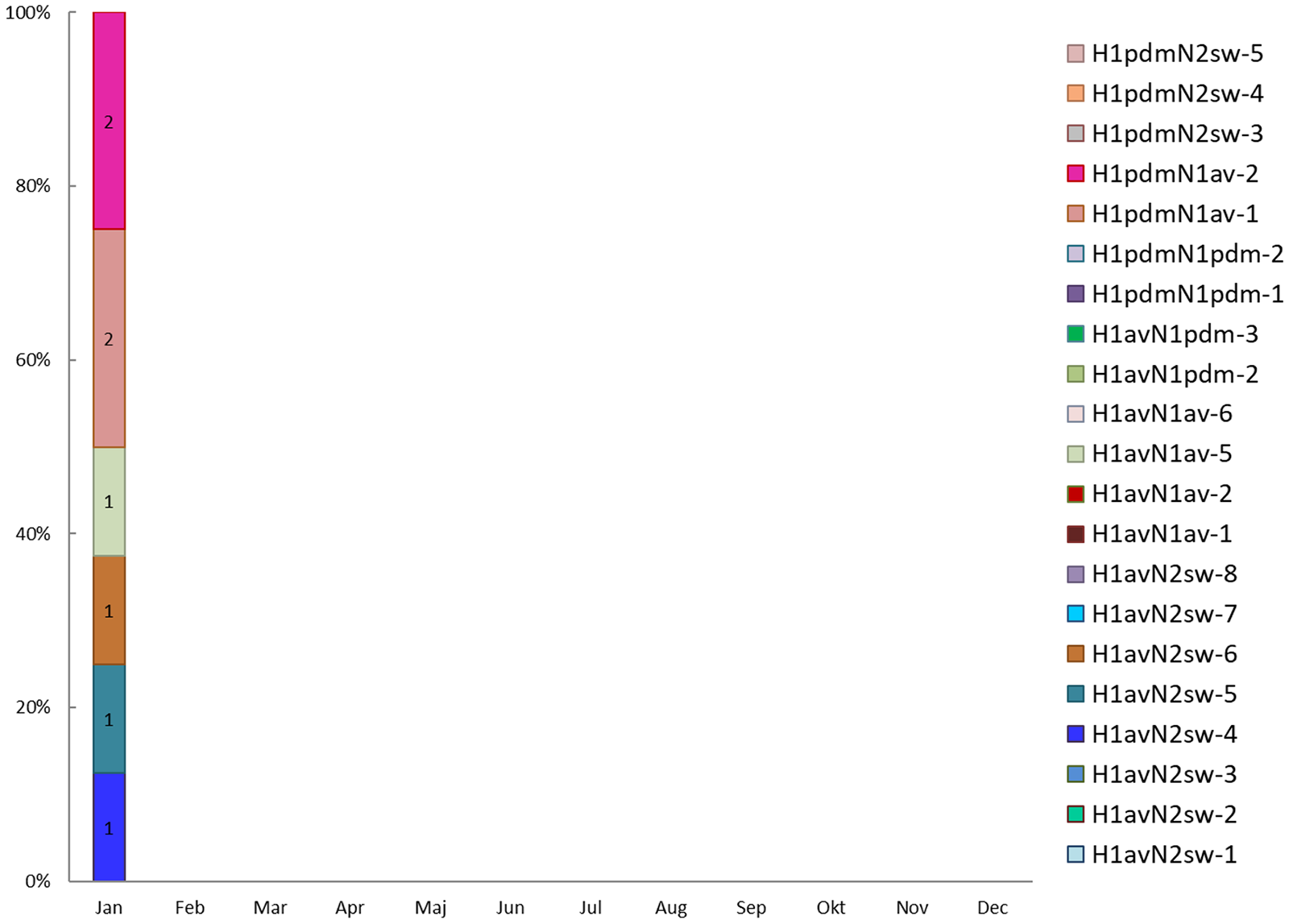
For December, 22 submissions were genotyped. In total eight different genotypes were observed, with H1avN2sw-5 being most often detected (n=6). In addition, the H1avN2sw-3 and H1pdmN1av-2 genotypes were also observed in four and three submissions respectively, both carrying an NS segment of Eurasian avian-like origin, whereas the remaining internal genes are of H1N1pdm09 origin.
Facts about the surveillance
DK-VET is responsible for a systematic prospective passive surveillance of circulating influenza A viruses in Danish swine herds based on samples obtained from pigs showing clinical signs of influenza. The overall aim of the surveillance is to acquire an overview on the contemporary circulating subtypes and genotypes in Danish pigs and in addition to aid in the diagnostics of disease causing agents among Danish pigs with the aim of securing the strategical purpose to reduce antibiotic use in Danish swine herds. Additionally, the results are used to evaluate both veterinary and zoonotic aspects of swine influenza A virus in Denmark.
Veterinary aspects
-
To obtain a better understanding of the complex epidemiology of swine influenza A virus in Denmark.
-
To enable establishment of virus stocks for fast vaccine production against novel subtypes causing enhanced disease in pigs.
-
To ensure that the applied diagnostic assays can detect all circulating swine influenza A viruses.
-
To document, especially for the Danish export of swine and pork, which influenza A virus subtypes that are present in Denmark.
-
To contribute to a common European overview on contemporary circulating swine influenza A virus subtypes.
Zoonotic aspects
-
Early detection and isolation of novel swine influenza A viruses having an increased zoonotic potential.
-
Early detection of molecular markers that have been related to an increased risk of swine-to-human transmission.
-
Early detection of viruses, containing genetic markers indicating that they are resistant for antiviral treatments.
-
Identification of genetic changes in the contemporary circulating swine influenza A virus strains that aids in improving diagnostic assays and protective vaccines, if transmission to humans occur.
The surveillance methodology
The surveillance is performed on samples from Danish swine herds submitting samples for diagnostic tests for swine influenza A virus at the Statens Serum Institut (SSI) or the Veterinary Laboratory, Kjellerup (Landbrug og Fødevarer) and includes:
-
Screening for influenza A virus using a pan-influenza A virus real time RT-PCR on diagnostic submissions for swine influenza A virus at the Staten Serum Institute (SSI) or the Veterinary Laboratory, Kjellerup (Landbrug og Fødevarer).
-
Screening of influenza A virus positive samples for pandemic influenza H1N1 (H1N1pdm09) by real time RT-PCR that specifically detects the HA gene segments in the H1N1pdm09 virus (H1pdm09).
-
Subtypning/genotypning by Next Generation Sequencing (NGS) on minimum one sample per influenza A virus positive submission.
Definitions and abbreviations
Influenza A virus has a RNA genome including eight segments, each containing minimum one gene that encodes for influenza virus proteins. In addition to the surface genes “hemagglutinin (HA)” and “neuraminidase (NA)” that determine the subtype, it is also important to consider the other ”internal genes”, as these play a role in the virulence and host specificity of the virus and define the genotype.
Below is a description of the swine influenza A virus subtypes and gene segments that so far have been detected in Danish pigs.
H1avN1av
”Avian-like” swine H1N1. This virus was introduced following an introduction of a full avian virus to swine from birds in the end of the seventies/early eighties. This virus is regarded as enzootic in Denmark.
H3swN2sw
”Swine H3N2”. This virus originates from the human pandemic in Hong Kong in 1968, which later adapted to swine by reassorting and obtaining an internal gene cassette of ”avian-like” H1N1 origin. This virus was detected with first time in Denmark in 1990 and has not been detected the last eight years in the surveillance.
H1avN2sw
This virus is also termed ”H1N2dk”, and it has the same ”avian-like” H1N1 HA gene and internal cassette as the ”avian-like” H1N1. However, the NA gene originates from H3swN2sw. This virus was detected for the first time in Denmark in 2003 and is considered enzootic in Denmark.
H1N1pdm09
This virus was the cause of the humane influenza pandemic in 2009 initiated in Mexico. All the gene segments of this virus are different from the earlier mentioned enzootic swine influenza A viruses in Denmark. This virus is now also considered enzootic in Denmark.
H1huN2sw
This virus was detected for the first time in England in 1994 and is a reassortant virus between a human seasonal influenza virus, circulating in the eighties and the H3swN2sw. This virus has never been detected in Denmark but is circulating in the many European countries.
H1pdm09
Virus with the specific HA gene segment of H1N1pdm09 origin.
N1pdm09
Virus with the specific NA gene segment of H1N1pdm09 origin.
H1av
Virus with the specific HA gene segments of ”avian-like” H1N1 (H1avN1av).
N1av
Virus with the specific NA gene segments of ”avian-like” H1N1 (H1avN1av).
N2sw
Virus with the specific NA gene of H3swN2sw and H1avN2sw origin.
N2hu#
NA gene segment with an origin in human seasonal H3N3 influenza. The ”#” indicates which season.
H3hu#
HA gene segment with an origin in human seasonal H3N3 influenza. The ”#” indicates which season.
Overview of genotypes
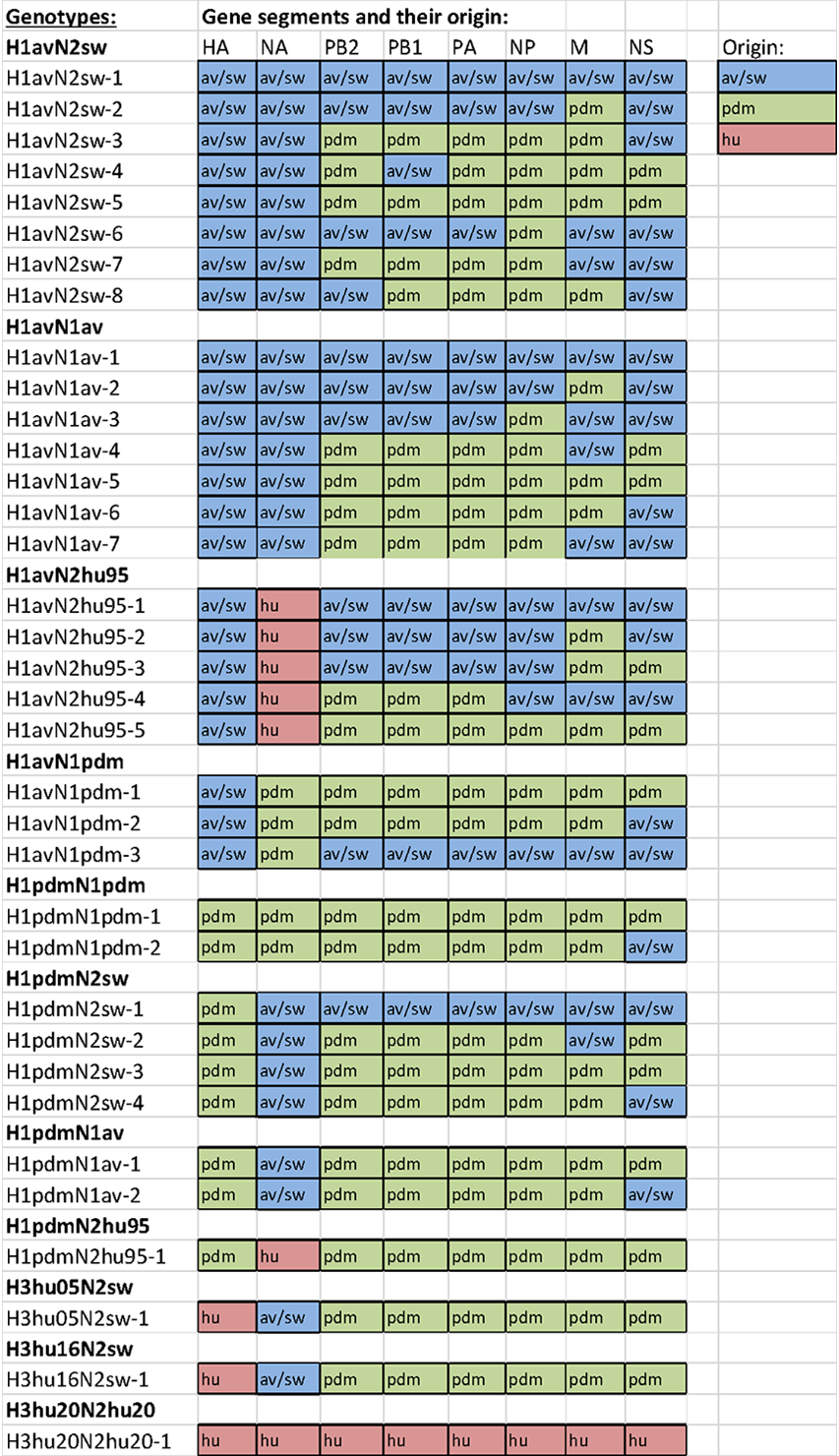
The eight gene segments of influenza A virus (HA, NA, PB2, PB1, PA, NP, M and NS) are listed on the top of the table. For each gene segment the origin is included by both color and letters. ”Av/sw” indicates avian/swine origin, ”pdm” indicates pandemic H1N1pdm09 origin and ”hu” indicates human seasonal origin. For each subtype a number of different genotypes is given, and represented by a numeral suffix.
Archive
2025
- Surveillance of Influenza A virus in Danish pigs, December 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, October 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, September 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, August 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, July 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, June 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, May 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, April 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, February 2025 (PDF)
- Surveillance of Influenza A virus in Danish pigs, January 2025 (PDF)
2024
- Overvågning af influenza A virus i svin, slutrapport 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, December 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, October 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, September 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, August 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, July 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, June 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, May 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, April 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, March 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, February 2024 (PDF)
- Surveillance of Influenza A virus in Danish pigs, January 2024 (PDF)
2023
- Overvågning af influenza A virus i svin, slutrapport 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, December 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, November 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, October 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, September 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, August 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, July 2023 (PDF)
- Surveillance of Influenza A virus in Danish pigs, June 2023 (PDF)
- Overvågning af Influenza A virus i svin, Maj 2023 (PDF)
- Overvågning af Influenza A virus i svin, April 2023 (PDF)
- Overvågning af Influenza A virus i svin, Marts 2023 (PDF)
- Overvågning af Influenza A virus i svin, Februar 2023 (PDF)
- Overvågning af Influenza A virus i svin, Januar 2023 (PDF)
2022
- Overvågning af influenza A virus i svin, slutrapport 2022 (PDF)
- Overvågning af Influenza A virus i svin, December 2022 (PDF)
- Overvågning af Influenza A virus i svin, November 2022 (PDF)
- Overvågning af Influenza A virus i svin, Oktober 2022 (PDF)
- Overvågning af Influenza A virus i svin, September 2022 (PDF)
- Overvågning af Influenza A virus i svin, August 2022 (PDF)
- Overvågning af Influenza A virus i svin, juni 2022 (PDF)
- Overvågning af Influenza A virus i svin, maj 2022 (PDF)
- Overvågning af Influenza A virus i svin, april 2022 (PDF)
- Overvågning af Influenza A virus i svin, marts 2022 (PDF)
- Overvågning af Influenza A virus i svin, februar 2022 (PDF)
- Overvågning af Influenza A virus i svin, januar 2022 (PDF)
2021
- Overvågning af influenza A virus i svin, 1. kvt. 2021 (PDF)
- Overvågning af influenza A virus i svin, 2. kvt. 2021 (PDF)
- Overvågning af influenza A virus i svin, 3. kvt. 2021 (PDF)
- Overvågning af influenza A virus i svin, 4. kvt. 2021 (PDF)
- Overvågning af influenza A virus i svin, slutrapport 2021 (PDF)
2020
- Overvågning af influenza A virus i svin, 1. kvt. 2020 (PDF)
- Overvågning af influenza A virus i svin, 2. kvt. 2020 (PDF)
- Overvågning af influenza A virus i svin, 3. kvt. 2020 (PDF)
- Overvågning af influenza A virus i svin, 4. kvt. 2020 (PDF)
- Overvågning af influenza A virus i svin, slutrapport 2020 (PDF)
2019